We may not see them, but microbes are all around. This fact is revealed to microbiology students who are tasked with a classic project: to identify bacteria and fungi from their environment. Armed with cotton swabs and Petri dishes full of nutient agar, students head out of the lab to see what lives on surfaces they encounter everyday.
Many students choose to sample the places they consider dirtiest: toilet handles, doorknobs, or the floors in the school hallway (when I took my first microbiology lab course, I sampled the dorm bathroom mirror). After swabbing and spreading the invisible contents onto the agar plate, we placed our agar plates in the incubator and awaited the microbial surprises the following class period.
You are viewing: Why Are Efforts Made To Identify Unknown Bacteria
Once the microbes revealed themselves on the agar plates, it was time to identify them. (To my disappointment, not much grew on my bathroom mirror plate. I later learned that despite the absence of microbes on my plate, there might still be microbes present on my bathroom mirror. Not all microbes grow on the same type of nutrients, or at the same temperature.)
This student project has many parallels to what microbiologists have been doing for centuries. From identifying microbes by physical and functional characteristics to the adaptation of more modern techniques, microbiologists (and future microbiologists) are continually building a vast toolkit to uncover the identities of previously unknown microscopic life.
Early Microbial Identification Studies By Microscopy
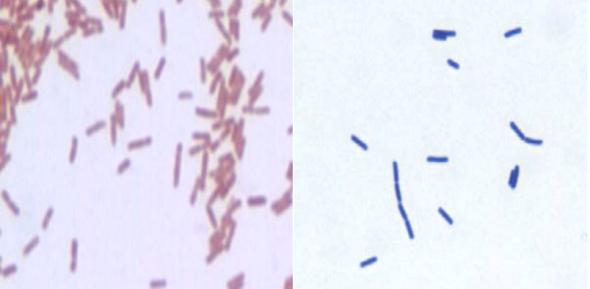
The earliest microbial identifications relied on observations of the microbe’s physical characteristics: shape, size, and the types of dyes it absorbed. Antoni van Leeuwenhoek first saw microbes through a microscope in the 1670s. These microbes came from decaying bodies, animals, vegetables, and water. He documented the findings, describing what he saw as “animalcules,” derived from the Latin “animalculum” or “tiny animal.”
To better visualize the microscopic amongst us, Hans Christian Gram developed the Gram stain technique in 1884. Gram created this technique to make bacteria more visible in stained lung tissue sections, and not for classifying microbes, as it is commonly applied today. Other types of staining can tell microbiologists whether certain features are present: spores (Schaeffer-Fulton staining), capsules (India ink or nigrosin) and mycolic acids (acid-fast staining).
How Agar Media Helps Us Identify Microbes
When scientists began cultivating microbes on agar media in the 1880s (thanks to the contributions of Angelina Hesse), they could more easily study the macroscopic characteristics of microbial populations. What color are the colonies? Do they look slimy? Or wrinkly? As microbiologists combined different formulations of nutrients with agar to grow a diverse set of microorganisms, they created another tool for microbial identification: selective and differential media that help microbiologists identify bacteria and yeast species. (Identifying viruses on agar plates is a different story and rely on methods such as differences in viral plaque phenotype.)
Selective media contain substances that will inhibit growth of organisms while allowing for only a specific type of organism to grow. For example, the high salt concentration in mannitol salt agar (MSA) inhibits the growth of most organisms except Staphylococcus species (thanks to brnQ, arsR, and cardiolipin). Selective media can also eliminate growth of specific organisms based on other criteria such as pH and amino acid composition.
Read more : Why Aren’t Birds Coming To My Feeder
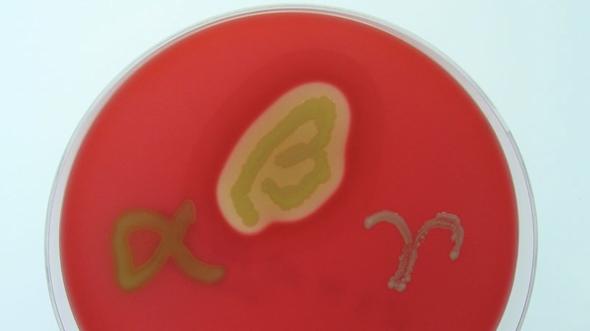
Differential media allow multiple bacterial species to grow but their growth patterns differ visually. Blood agar is a commonly used differential medium, containing 5-10% sheep or horse blood, a requirement for Streptococcus species to grow. Different Streptococcus species break down the blood cells (in a process called hemolysis) in different ways, leading to differences in appearance:
- No media color change = no blood cell lysis (S. veridans)
- Green/brown media = partial blood cell lysis (S. haemolyticus)
- Lightened agar around bacterial growth = complete blood cell lysis (S. pyogenes)
By combining different indicators and compounds into the same agar media formulation, media can be both selective and differential. The MSA media described above actually contains both selective (salt) and differential (mannitol) components. While salt selects for Staphylococcus species, the media also differentiates Staphylococcus species that ferment mannitol from those that don’t. If a bacterium fermented mannitol (e.g., S. aureus), it lowers the pH of the medium. The pH change is detectable because the media contains phenol red which turns yellow at low pH. If a bacterium does not ferment mannitol (e.g., S. epidermidis), the pH doesn’t lower and the medium remains red.
A similar example of media that is both differentiating and selecting is MacConkey Agar. This type of agar includes bile salts, which are found in the gut and help in digestion by emulsifying fats. Microorganisms that live in the intestines (called enteric microbes) constantly encounter bile salts, and have developed mechanisms to prevent these salts from destroying their membranes. Non-enteric microbes are more susceptible to bile salts and less likely to grow in their presence. Therefore, MacConkey Agar selects for bile-resistant microorganisms. The current recipe of MacConkey Agar contains 2 extra ingredients that increase its selectivity, and make it differential: (1) the addition of crystal violet to the MacConkey agar recipe inhibits growth of Gram-positive organisms, and (2) the addition of a pH indicator, neutral red, differentiate lactose fermenters from non-fermenters.
While these are just a few examples of how types of media can help microbiologists distinguish between microbes, there are many other types of selective and differential media. For example:
- Selective media:
- YM agar selects for microbes that grow in low pH conditions such as yeasts and molds.
- Eosin methylene blue selects for Gram-negative organisms.
- Baird-Parker selects for Gram-positive Staphylococci.
- Buffered charcoal yeast extract agar selects for some Gram-negatives, especially Legionella pneumophila.
- Sarbouraud’s agar, which has a low pH and high glucose concentration, selects for some fungi.
- Differential media:
- X-gal plates identifies lac operon mutants during clone selection.
- Eosin methylene blue differentiates between lactose fermenters and non-fermenters.
Biochemical Tests Used to Identify Microbes
Microbiologists can ask additional questions about microbial identity based on microbial behavior during biochemical tests.
Some biochemical tests for microbial identification are quite simple. To test whether bacteria contain a catalase enzyme, a microbiologist drops hydrogen peroxide into a smear of bacteria on a microscope slide. If the bacteria contain catalase, the mixture bubbles as the hydrogen peroxide decomposes into water and oxygen. In the clinic, the catalase test helps distinguish catalase-positive Staphylococci from catalase-negative Streptococcus, which are both Gram-positive cocci.
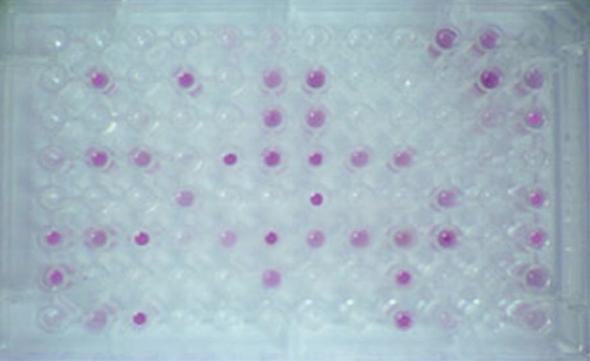
In substrate utilization tests, a panel of substrates, such as carbon or nitrogen sources, can quickly test a microbe’s ability to use different substrates at the same time. These assays are typically done in 96 well plates where each well contains a different substrate. If a microbe can use the given substrate, it will generate a color change in the medium. By analyzing the combination of substrates utilized on the plate, the bacteria in question could be identified.
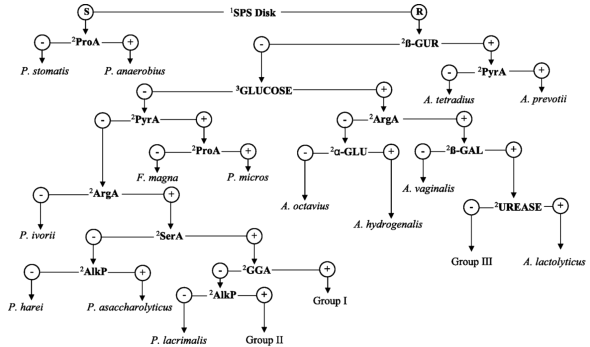
Given the wealth of agar media, microscopy stains, and biochemical tests, microbiologists have built flow charts to identity the bacteria surrounding us. Yet, the numerous growth and biochemical tests that microbiologists have amassed cannot precisely reveal all of the ways one microbe may be different from another. These tests also require that the microbes in question be culturable. How do microbiologists identify the many microbes that are unculturable?
What Does the DNA Say? 16S rRNA and Whole Genome Sequencing
Read more : Why Was Opie’s Mom Never Mentioned
DNA sequencing ushered in many newer techniques to identify microbes more precisely, while simultaneously providing information about microbial function. Unlike the methods described above, sequencing does not require the microbiologist to first grow the organism.
One of these first DNA sequencing methods is 16S rRNA gene sequencing and relies on the fact that the 16S rRNA is a relatively stable region with a slow rate of evolution. This makes the sequence a great interrogation point to determine relationships between species. The logic is if organisms are closely related, their 16S rRNA gene sequences will be more similar than organisms that are not closely related. To sequence the 16S rRNA gene, you’d first have to amplify the region by PCR and then sequence the product. However, this provides a small piece of the microbial puzzle.
Sequencing all of the DNA in a microbe and assembling these sequences into a genome reveals much more than 16S rRNA gene sequencing can. You can get information about nearly all of the genes in the organism and get a sense of what the microbe is capable of doing. There are several methods of DNA sequencing used to generate a whole genome sequence.
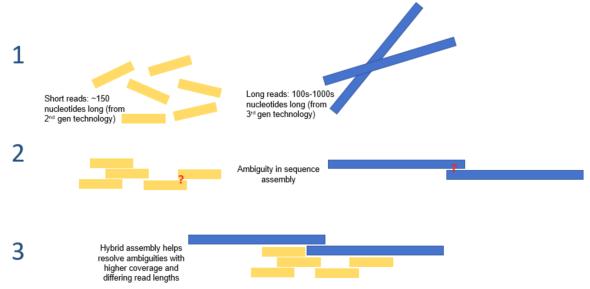
In next generation sequencing (NGS), or massively parallel sequencing, genomic DNA is broken into small segments that are sequenced simultaneously. This results in 1 million to 43 billion short reads (50-400 bp) per run. The large number of reads can be assembled into longer fragments, but often times does not result in complete assembly of a genome.
Long-read sequencing methods read over 10 kb at once. One popular long-read method is Nanopore sequencing; here, a single-stranded DNA molecule is fed through a very small pore (hence the name Nanopore). As the DNA strand passes through the pore, the surrounding electrical field changes in ways specific to the DNA sequence in the pore. By observing changes in the current, the DNA sequence can be inferred as the molecule passes through the nano pore.
Long-read sequencing and NGS can be paired together in a method called hybrid assembly. When paired with long-read sequence, any ambiguity that exists in NGS results can be better deciphered, and vice versa.
Sequencing methods for microbial identification have some additional advantages over media-based methods and biochemical tests. While the agar media-based methods and biochemical tests are used for identifying bacteria and fungi, they aren’t developed for identifying virus and can only be used for organisms that are culturable. Sequencing provides a more robust toolset, since it can identify both viruses and unculturable microbes.
Microbial Identification: Beyond the Classroom
The microbial identification project common in many microbiology lab course reminds us that microbes are all around. But despite the number of bacteria and fungi that grow from swabbed phones or water bottles, the majority are not harmful. Microbial identification is not just limited to the classroom however. In clinical labs, microbiologists identify the microbes behind disease in their patients. And for basic research, microbiologists all over the world are studying where microbes reside and what they are doing: sourdough starters, showerheads, the subway, oceans, and soils are just the beginnings of our microbial exploration.
The above represent the views of the author and does not necessarily reflect the opinion of the American Society for Microbiology.
Source: https://t-tees.com
Category: WHY
